Summary
Clan
This family is a member of clan (CL00066), which contains the following 5 members:
sn2903 snoMe28S-G3255 snoR1 SNORD60 snosnR48Wikipedia annotation Edit Wikipedia article
The Rfam group coordinates the annotation of Rfam families in Wikipedia. This family is described by a Wikipedia entry Small nucleolar RNA Me28S-Gm3255. More...
This page is based on a Wikipedia article. The text is available under the Creative Commons Attribution/Share-Alike License.
Sequences
Quick actions
Table view (49 sequence regions)
| Original order | Download FASTA | Accession | Bit score | Type | Start | End | Description | Species | View context |
|---|---|---|---|---|---|---|---|---|---|
| 0 | AC008186.5 | N/A | seed | 23,701 | 23,785 | Drosophila melanogaster, chromosome 2R, region 45A-46A, BAC clone BACR10M14, complete sequence. | Drosophila melanogaster (fruit fly) |
|
|
| 1 | AC008186.5 | N/A | seed | 24,468 | 24,549 | Drosophila melanogaster, chromosome 2R, region 45A-46A, BAC clone BACR10M14, complete sequence. | Drosophila melanogaster (fruit fly) |
|
|
| 2 | AAPP01018724.1 | N/A | seed | 106,634 | 106,550 | Drosophila ananassae strain TSC#14024-0371.13 Ctg01_18746, whole genome shotgun sequence. | Drosophila ananassae |
|
|
| 3 | AAEU02000094.1 | N/A | seed | 193,595 | 193,512 | Drosophila yakuba strain Tai18E2 chromosome 2L v2_Cont1.39, whole genome shotgun sequence. | Drosophila yakuba |
|
|
| 4 | AAFS01000046.1 | N/A | seed | 161,956 | 162,039 | Drosophila pseudoobscura pseudoobscura strain MV2-25 3207210, whole genome shotgun sequence. | Drosophila pseudoobscura pseudoobscura |
|
|
| 5 | AAGH01023660.1 | N/A | seed | 5,285 | 5,360 | Drosophila simulans strain white501 chromosome 2R Cont36.73, whole genome shotgun sequence. | Drosophila simulans |
|
|
| 6 | AAPQ01007360.1 | N/A | seed | 2,428,596 | 2,428,511 | Drosophila erecta strain TSC#14021-0224.01 Ctg01_7359, whole genome shotgun sequence. | Drosophila erecta |
|
|
| 7 | AASS01036184.1 | N/A | seed | 1,711 | 1,633 | Drosophila simulans strain MD106TS Cont471.2, whole genome shotgun sequence. | Drosophila simulans |
|
|
| 8 | AAKO01000502.1 | N/A | seed | 98,060 | 98,140 | Drosophila sechellia strain Rob3c cont1.000501, whole genome shotgun sequence. | Drosophila sechellia |
|
|
| 9 | AAEU02000094.1 | N/A | seed | 192,782 | 192,704 | Drosophila yakuba strain Tai18E2 chromosome 2L v2_Cont1.39, whole genome shotgun sequence. | Drosophila yakuba |
|
|
| 0 | AE013599.5 | 100.40 | full | 9,096,763 | 9,096,844 | Drosophila melanogaster chromosome 2R. | Drosophila melanogaster (fruit fly) |
|
|
| 1 | CM000164.1 | 93.70 | full | 3,752,062 | 3,752,140 | CM000164.1 Drosophila simulans strain white501 chromosome 2R, whole genome shotgun sequence | Drosophila simulans |
|
|
| 2 | CH480816.1 | 93.40 | full | 2,622,516 | 2,622,596 | Drosophila sechellia strain Rob3c scaffold_1 genomic scaffold, whole genome shotgun sequence. | Drosophila sechellia |
|
|
| 3 | NW_016055718.1 | 90.90 | full | 81,653 | 81,733 | NW_016055718.1 Drosophila rhopaloa unplaced genomic scaffold, Drho_2.0 scf7180000773652, whole genome shotgun sequence | Drosophila rhopaloa |
|
|
| 4 | CH479183.1 | 87.20 | full | 5,021,081 | 5,021,164 | CH479183.1 Drosophila persimilis strain MSH-3 scaffold_4 genomic scaffold, whole genome shotgun sequence | Drosophila persimilis |
|
|
| 5 | NC_046680.1 | 87.20 | full | 13,431,226 | 13,431,309 | NC_046680.1 Drosophila pseudoobscura strain MV2-25 chromosome 3, UCI_Dpse_MV25, whole genome shotgun sequence | Drosophila pseudoobscura pseudoobscura |
|
|
| 6 | CH902619.1 | 86.30 | full | 3,513,783 | 3,513,699 | CH902619.1 Drosophila ananassae strain TSC#14024-0371.13 scaffold_13266 genomic scaffold, whole genome shotgun sequence | Drosophila ananassae |
|
|
| 7 | CH480816.1 | 85.80 | full | 2,621,746 | 2,621,830 | Drosophila sechellia strain Rob3c scaffold_1 genomic scaffold, whole genome shotgun sequence. | Drosophila sechellia |
|
|
| 8 | CM000164.1 | 85.80 | full | 3,751,286 | 3,751,370 | CM000164.1 Drosophila simulans strain white501 chromosome 2R, whole genome shotgun sequence | Drosophila simulans |
|
|
| 9 | NW_016067408.1 | 84.80 | full | 2,424,060 | 2,423,977 | NW_016067408.1 Drosophila kikkawai unplaced genomic scaffold, Dkik_2.0 scf7180000302476, whole genome shotgun sequence | Drosophila kikkawai |
|
|
| 10 | AE013599.5 | 82.80 | full | 9,095,996 | 9,096,080 | Drosophila melanogaster chromosome 2R. | Drosophila melanogaster (fruit fly) |
|
|
| 11 | JAMKOV010000007.1 | 82.40 | full | 852,118 | 852,196 | JAMKOV010000007.1 Drosophila gunungcola strain Sukarami chromosome 2R 000006F, whole genome shotgun sequence | Drosophila gunungcola |
|
|
| 12 | NW_016055718.1 | 80.80 | full | 80,788 | 80,872 | NW_016055718.1 Drosophila rhopaloa unplaced genomic scaffold, Drho_2.0 scf7180000773652, whole genome shotgun sequence | Drosophila rhopaloa |
|
|
| 13 | JAMKOV010000007.1 | 79.90 | full | 851,279 | 851,363 | JAMKOV010000007.1 Drosophila gunungcola strain Sukarami chromosome 2R 000006F, whole genome shotgun sequence | Drosophila gunungcola |
|
|
| 14 | NW_016067408.1 | 78.90 | full | 2,424,301 | 2,424,216 | NW_016067408.1 Drosophila kikkawai unplaced genomic scaffold, Dkik_2.0 scf7180000302476, whole genome shotgun sequence | Drosophila kikkawai |
|
|
| 15 | CH479183.1 | 73.20 | full | 5,020,795 | 5,020,876 | CH479183.1 Drosophila persimilis strain MSH-3 scaffold_4 genomic scaffold, whole genome shotgun sequence | Drosophila persimilis |
|
|
| 16 | NC_046680.1 | 72.10 | full | 13,430,941 | 13,431,021 | NC_046680.1 Drosophila pseudoobscura strain MV2-25 chromosome 3, UCI_Dpse_MV25, whole genome shotgun sequence | Drosophila pseudoobscura pseudoobscura |
|
|
| 17 | NC_047629.2 | 59.60 | full | 16,989,667 | 16,989,749 | NC_047629.2 Drosophila albomicans strain 15112-1751.03 chromosome 3, ASM965048v2, whole genome shotgun sequence | Drosophila albomicans |
|
|
| 18 | CH964282.1 | 56.40 | full | 3,773,231 | 3,773,312 | CH964282.1 Drosophila willistoni strain TSC#14030-0811.24 scf2_1100000004954 genomic scaffold, whole genome shotgun sequence | Drosophila willistoni |
|
|
| 19 | LR899009.1 | 56.10 | full | 30,544,537 | 30,544,619 | LR899009.1 Hermetia illucens genome assembly, chromosome: 1 | Hermetia illucens |
|
|
| 20 | WJQU01001944.1 | 55.50 | full | 23,365 | 23,447 | WJQU01001944.1 Pseudolycoriella hygida isolate HSMRA1968 HiC_scaffold_1944, whole genome shotgun sequence | Bradysia hygida |
|
|
| 21 | CAJVCH010250071.1 | 51.00 | full | 129 | 49 | CAJVCH010250071.1 Allacma fusca genome assembly, contig: NODE_327101, whole genome shotgun sequence | Allacma fusca |
|
|
| 22 | JACASF010000005.1 | 50.50 | full | 44,996,767 | 44,996,838 | JACASF010000005.1 Molossus molossus isolate mMolMol1 scaffold_m16_p_5, whole genome shotgun sequence | Molossus molossus (Pallas's mastiff bat) |
|
|
| 23 | CP012524.1 | 50.50 | full | 7,566,245 | 7,566,164 | Drosophila busckii chromosome 2R sequence. | Drosophila busckii |
|
|
| 24 | NW_022060529.1 | 49.70 | full | 21,960,010 | 21,960,089 | NW_022060529.1 Scaptodrosophila lebanonensis strain 11010-0011.00,11010-0021.00 unplaced genomic scaffold, SlebRS2 tig00000001-arrow-pilon, whole genome shotgun sequence | Scaptodrosophila lebanonensis |
|
|
| 25 | LSRL02000007.1 | 49.40 | full | 310,167 | 310,244 | LSRL02000007.1 Drosophila navojoa isolate navoj_Jal97 7, whole genome shotgun sequence | Drosophila navojoa |
|
|
| 26 | LJIJ01000034.1 | 49.00 | full | 355,174 | 355,254 | Orchesella cincta Ocin01_Sc0034, whole genome shotgun sequence. | Orchesella cincta |
|
|
| 27 | CAJVCH010253372.1 | 48.50 | full | 129 | 49 | CAJVCH010253372.1 Allacma fusca genome assembly, contig: NODE_330105, whole genome shotgun sequence | Allacma fusca |
|
|
| 28 | CH933808.1 | 48.30 | full | 5,124,513 | 5,124,589 | CH933808.1 Drosophila mojavensis strain TSC#15081-1352.22 scaffold_6496 genomic scaffold, whole genome shotgun sequence | Drosophila mojavensis |
|
|
| 29 | NCKV01000766.1 | 47.80 | full | 6,435 | 6,514 | NCKV01000766.1 Leptotrombidium deliense isolate UoL-UT scaffold0000000767, whole genome shotgun sequence | Leptotrombidium deliense |
|
|
| 30 | CM019174.2 | 47.10 | full | 12,822,685 | 12,822,607 | CM019174.2 Apolygus lucorum isolate 12Hb linkage group LG16, whole genome shotgun sequence | Apolygus lucorum |
|
|
| 31 | CM019174.2 | 47.00 | full | 12,833,888 | 12,833,810 | CM019174.2 Apolygus lucorum isolate 12Hb linkage group LG16, whole genome shotgun sequence | Apolygus lucorum |
|
|
| 32 | CH940648.1 | 46.20 | full | 367,281 | 367,201 | CH940648.1 Drosophila virilis strain TSC#15010-1051.87 scaffold_12875 genomic scaffold, whole genome shotgun sequence | Drosophila virilis |
|
|
| 33 | CM019174.2 | 45.90 | full | 12,822,493 | 12,822,415 | CM019174.2 Apolygus lucorum isolate 12Hb linkage group LG16, whole genome shotgun sequence | Apolygus lucorum |
|
|
| 34 | CAJHJT010000056.1 | 45.50 | full | 37,916,990 | 37,916,913 | CAJHJT010000056.1 Ceratitis capitata strain EGII genome assembly, contig: scaffold_6, whole genome shotgun sequence | Ceratitis capitata (Mediterranean fruit fly) |
|
|
| 35 | NW_015381949.1 | 45.40 | full | 191,710 | 191,790 | NW_015381949.1 Neodiprion lecontei isolate Nlec_01 unplaced genomic scaffold, Nlec1.0, whole genome shotgun sequence | Neodiprion lecontei |
|
|
| 36 | CM019174.2 | 45.40 | full | 12,822,263 | 12,822,185 | CM019174.2 Apolygus lucorum isolate 12Hb linkage group LG16, whole genome shotgun sequence | Apolygus lucorum |
|
|
| 37 | ADMH02000483.1 | 45.10 | full | 92,906 | 92,825 | Anopheles darlingi Cont2812, whole genome shotgun sequence. | Anopheles darlingi |
|
|
| 38 | CH964282.1 | 44.30 | full | 3,774,143 | 3,774,224 | CH964282.1 Drosophila willistoni strain TSC#14030-0811.24 scf2_1100000004954 genomic scaffold, whole genome shotgun sequence | Drosophila willistoni |
|
|
| 39 | OV725079.1 | 44.30 | full | 44,068,252 | 44,068,163 | OV725079.1 Nezara viridula genome assembly, chromosome: 3 | Nezara viridula (southern green stink bug) |
|
Alignment
There are various ways to view or download the seed alignments that we store. You can use a sequence viewer to look at them, or you can look at a plain text version of the sequence in a variety of different formats. More...
View options
You can view Rfam seed alignments in your browser in various ways. Choose the viewer that you want to use and click the "View" button to show the alignment in a pop-up window.
Formatting options
You can view or download Rfam seed alignments in several formats. Check either the "download" button, to save the formatted alignment, or "view", to see it in your browser window, and click "Generate".
Download
Download a gzip-compressed, Stockholm-format file containing the seed alignment for this family. You may find RALEE useful when viewing sequence alignments.
Submit a new alignment
We're happy receive updated seed alignments for new or existing families. Submit your new alignment and we'll take a look.
Secondary structure
Species distribution
Sunburst controls
HideLineage
Move your mouse over the main tree to show the lineage of a particular node.
You can move this pane by dragging it.
Weight segments by...
Change the size of the sunburst
Colour assignments
 Archea
Archea
|
 Eukaryota
Eukaryota
|
 Bacteria
Bacteria
|
 Other sequences
Other sequences
|
 Viruses
Viruses
|
 Unclassified
Unclassified
|
 Viroids
Viroids
|
 Unclassified sequence
Unclassified sequence
|
Selections
Click on a node to select that node and its sub-tree.
Clear selection
This visualisation provides a simple graphical representation of the distribution of this family across species. You can find the original interactive tree in the adjacent tab. More...
Tree controls
HideThe tree shows the occurrence of this RNA across different species. More...
Loading...
Please note: for large trees this can take some time. While the tree is loading, you can safely switch away from this tab but if you browse away from the family page entirely, the tree will not be loaded.
Trees
This page displays the predicted phylogenetic tree for the alignment. More...
Note: You can also download the data file for the seed tree.
References
This section shows the database cross-references that we have for this Rfam family.
Literature references
-
Huang ZP, Zhou H, He HL, Chen CL, Liang D, Qu LH RNA. 2005;11:1303-1316. Genome-wide analyses of two families of snoRNA genes from Drosophila melanogaster, demonstrating the extensive utilization of introns for coding of snoRNAs. PUBMED:15987805
-
Kiss T; Cell 2002;109:145-148. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. PUBMED:12007400
External database links
| Gene Ontology: | GO:0006396 (RNA processing); GO:0005730 (nucleolus); |
| Sequence Ontology: | SO:0000593 (C_D_box_snoRNA); |
Curation and family details
This section shows the detailed information about the Rfam family. We're happy to receive updated or improved alignments for new or existing families. Submit your new alignment and we'll take a look.
Curation
| Seed source | PMID:15987805 | ||||||
| Structure source | Predicted; PFOLD; Moxon SJ, Daub J | ||||||
| Type | Gene; snRNA; snoRNA; CD-box; | ||||||
| Author |
Moxon SJ
|
||||||
| Alignment details |
|
Model information
| Build commands |
cmbuild -F CM SEED
cmcalibrate --mpi CM
|
| Search command |
cmsearch --cpu 4 --verbose --nohmmonly -T 38.93 -Z 2958934 CM SEQDB
|
| Gathering cutoff | 44.0 |
| Trusted cutoff | 44.3 |
| Noise cutoff | 43.8 |
| Covariance model | Download |
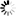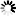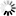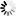 Loading...
Loading...